Genetic Fine Structure or Fine
Structure of Gene

A gene (Figur) is a
locatable region of genomic sequence, corresponding to a unit
of inheritance, which is
associated with regulatory regions, transcribed regions and/or other functional
sequence regions. The physical development and phenotype
of organisms can be thought of as a product of genes interacting with each
other and with the environment. A concise definition of a gene, taking into
account complex patterns of regulation and transcription, genic conservation
and non-coding RNA genes, has been proposed by Gerstein et al. "A gene is
a union of genomic sequences encoding a coherent set of potentially overlapping
functional products".
Colloquially, the term gene is often used to refer to an
inheritable trait which is usually accompanied by a phenotype as in ("tall
genes" or "bad genes") -- the proper scientific term for this is
allele.
In cells, genes consist of a long strand of DNA that contains a promoter,
which controls the activity of a gene, and coding and non-coding sequence.
Coding sequence determines what the gene produces, while non-coding sequence
can regulate the conditions of gene expression.
When a gene is active, the coding and non-coding sequence is copied in a
process called transcription, producing an RNA copy of the gene's
information. This RNA can then direct the synthesis of proteins
via the genetic code.
But some RNAs are used directly, for example as part of the ribosome.
These molecules resulting from gene expression, whether RNA or protein, are known
as gene products.
Genes often contain regions that do not encode products,
but regulate gene expression. The genes of eukaryotic
organisms can contain regions called introns that are
removed from the messenger RNA in a process called splicing. The regions encoding gene products
are called exons.
In eukaryotes, a single gene can encode multiple proteins, which are produced
through the creation of different arrangements of exons through alternative splicing. In prokaryotes
(bacteria
and archaea),
introns are less common and genes often contain a single uninterrupted stretch
of DNA, called a cistron, that codes for a product. Prokaryotic genes are often
arranged in groups called operons with promoter and operator sequences that regulate transcription of a single long RNA. This RNA contains
multiple coding sequences. Each coding sequence is preceded by a Shine-Dalgarno sequence that ribosomes
recognize.
The total set of genes in an organism is known as its genome. An
organism's genome size is generally lower in prokaryotes,
both in number of base pairs and number of genes, than even single-celled eukaryotes.
However, there is no clear relationship between genome sizes and complexity in
eukaryotic organisms.
One of the largest known genomes belongs to the single-celled
amoeba Amoeba dubia, with over 670 billion base
pairs, some 200 times larger than the human genome. The estimated number of
genes in the human genome has been repeatedly revised downward since the
completion of the Human Genome Project; current estimates place
the human genome at just under 3 billion base pairs and about 20,000–25,000
genes. A recent Science article gives a number of 20,488
protein-coding genes, with perhaps 100 more yet to be discovered. The gene
density of a genome is a measure of the number of genes per million base pairs
(called a Megabase,
Mb); prokaryotic genomes have much higher gene densities than eukaryotes. The
gene density of the human genome is roughly 12–15 genes per megabase pair.
History
The existence of genes was first suggested by Gregor Mendel
(1822-1884), who, in the 1860s, studied inheritance in pea plants.
Mendel's concept was given a name by Hugo de Vries
in 1889, who, at that time probably unaware of Mendel's work, in his book
Intracellular Pangenesis coined the term "pangen" for "the
smallest particle [representing] one hereditary characteristic".
Wilhelm Johannsen abbreviated this term to
"gene" ("gen" in Danish and German) two decades later.
Physical Structure –
The vast majority of living
organisms encode their genes in long strands of DNA. DNA consists of a
chain made from four types of nucleotide subunits: adenine,
cytosine,
guanine,
and thymine
(Figure ).

Each nucleotide subunit consists of three components: a phosphate group, a deoxyribose sugar ring, and a nucleobase. Thus, nucleotides in DNA or RNA are typically called 'bases'; consequently they are commonly referred to simply by their purine or pyrimidine original base components adenine, cytosine, guanine, thymine.
Adenine and
guanine are purines and cytosine and thymine are pyrimidines. The most common
form of DNA in a cell is in a double helix
structure, in which two individual DNA strands twist around each other in a
right-handed spiral. In this structure, the base pairing rules specify that guanine
pairs with cytosine
and adenine
pairs with thymine
(each pair contains one purine and one pyrimidine). The base pairing between
guanine and cytosine forms three hydrogen bonds, while the base pairing between
adenine and thymine forms two hydrogen bonds. The two strands in a double helix
must therefore be complementary, that is, their bases must align such that the
adenines of one strand are paired with the thymines of the other strand, and so
on.
Due to the chemical composition of the pentose residues of
the bases, DNA strands have directionality. One end of a DNA polymer contains
an exposed hydroxyl
group on the deoxyribose, this is known as the 3' end
of the molecule. The other end contains an exposed phosphate
group, this is the 5' end.
The directionality of DNA is vitally important to many cellular processes,
since double helices are necessarily directional (a strand running 5'-3' pairs
with a complementary strand running 3'-5') and processes such as DNA replication
occur in only one direction. All nucleic acid synthesis in a cell occurs in the
5'-3' direction, because new monomers are added via a dehydration
reaction that uses the exposed 3' hydroxyl as a nucleophile.
The expression of genes encoded in DNA begins by transcribing the gene into RNA, a second type of nucleic acid
that is very similar to DNA, but whose monomers contain the sugar ribose rather than deoxyribose.
RNA also contains the base uracil
in place of thymine.
RNA molecules are less stable than DNA and are typically single-stranded. Genes
that encode proteins
are composed of a series of three-nucleotide
sequences called codons,
which serve as the "words" in the genetic "language". The genetic code
specifies the correspondence during protein translation between codons and amino acids.
The genetic code is nearly the same for all known organisms.
RNA genes –
In some cases, RNA is an intermediate
product in the process of manufacturing proteins from genes. However, for other
gene sequences, the RNA molecules are the actual functional products. For
example, RNAs known as ribozymes are capable of enzymatic function,
and miRNAs
have a regulatory role. The DNA
sequences from which such RNAs are transcribed are known as RNA genes.
Some viruses
store their entire genomes in the form of RNA, and contain no DNA at all.
Because they use RNA to store genes, their cellular
hosts
may synthesize their proteins as soon as they are infected
and without the delay in waiting for transcription. On the other hand, RNA retroviruses,
such as HIV,
require the reverse transcription of their genome from RNA into
DNA before their proteins can be synthesized. In 2006, French researchers came
across a puzzling example of RNA-mediated inheritance in mouse. Mice with a loss-of-function mutation in the gene Kit have white tails.
Offspring of these mutants can have white tails despite having only normal Kit
genes. The research team traced this effect back to mutated Kit RNA. While RNA
is common as genetic storage material in viruses, in mammals in particular RNA
inheritance has been observed very rarely.
Functional structure of a gene –
All genes have regulatory regions in addition to regions that explicitly code for a protein or RNA product. A regulatory region shared by almost all genes is known as the promoter (Figure ),
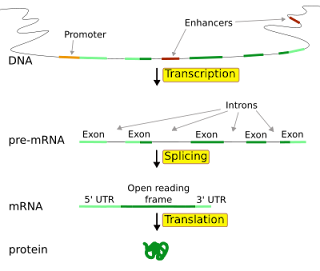
which provides a position that is recognized by
the transcription machinery when a gene is about to be transcribed and
expressed.
Although promoter regions have a consensus sequence that is the most common sequence at this position, some genes have "strong" promoters that bind the transcription mach-inery well, and others have "weak" promoters that bind poorly. These weak promoters usually permit a lower rate of transcription than the strong promote-rs, because the transcription mach-inery binds to them and initiates trans-cripttion less frequently. Other possible regulatory regions include enhancers, which can compensate for a weak promoter. Most regulatory regions are "upstream" - that is, before or toward the 5' end of the transcription initiation site. Eukaryotic promoter regions are much more complex and difficult to identify than prokaryotic promoters.
Many prokaryotic genes are organized into operons, or groups
of genes whose products have related functions and which are transcribed as a
unit. By contrast, eukaryotic genes are transcribed only one at a
time, but may include long stretches of DNA called introns which are
transcribed but never translated into protein (they are spliced out before
translation). Splicing can also occur in prokaryotic genes, but is less common
than in eukaryotes.
Comments
Post a Comment