Phage Phenotypes
Bacteriophage - A bacteriophage (from 'bacteria'
and Greek
phagein, 'to eat') is any one of a number of viruses
that infect
bacteria. The term is commonly used in its shortened form, phage.
Typically, bacteriophages consist of an outer protein
hull enclosing genetic material. The genetic material can be ssRNA (single stranded RNA), dsRNA, ssDNA, or dsDNA between 5 and 500
kilo base pairs
long with either circular or linear arrangement. Bacteriophages are much
smaller than the bacteria they destroy - usually between 20 and 200 nm in
size.
T2 and
its close relative T4 are
viruses that infect the bacterium E. coli. The infection ends with
destruction (lysis) of the bacterial cell so these viruses are examples of
bacteriophages ("bacteria eaters").
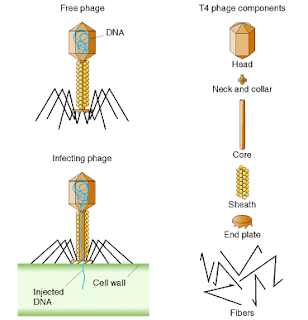
General Phenotype - Generally
each virus particle (virion) consists of:
- a protein head (~0.1 µm) inside of which is a single,
circular molecule of double-stranded DNA containing 166,000 base pairs.
(Figure 2.1)
- a protein tail from which extend thin protein fibers
Life
Cycle - The virus attaches to the E. coli cell. This requires a precise molecular interaction between
the fibers and the cell wall of the host. The DNA molecule is injected into the
cell. Within 1 minute, the viral DNA begins to be transcribed
and translated
into some of the viral proteins, and synthesis of host proteins is stopped. At
5 minutes, viral enzymes needed for synthesis of new viral DNA molecules are
produced. At 8 minutes, some 40 different structural proteins for the viral
head and tail are synthesized. At 13 minutes, assembly of new viral particles
begins. At 25 minutes, the viral lysozyme
destroys the bacterial cell wall and the viruses burst out — ready to infect
new hosts.
- If the bacterial cells
are growing in liquid culture, it turns clear.
- If the bacterial cells
are growing in a "lawn" on the surface of an agar plate, then
holes, called plaques
(Figure 2.2), appear in the lawn.
New Phenotypes - Occasionally,
new phenotypes appear such as a change in the appearance of the plaques or even
a loss in the ability to infect the host.
Examples:
- h
o Some strains of E.
coli, e.g. one designated B/2, gain the ability to resist infection by
normal ("wild-type") T2. The mutation has caused a change in the
structure of their cell wall so that the tail fibers of T2 can no longer bind
to it. However, T2 can strike back. Occasional T2 mutants appear that overcome
this resistance. The mutated gene, designated h (for "host range"),
encodes a change in the tail fibers so they can once again bind to the cell
wall of strain B/2. The normal of "wild-type" gene is designated h+
.
o When plated on a lawn containing both E. coli B and E. coli B/2,
§ the mutant (h) viruses can lyze both strains of E. coli, producing clear plaques, while
§ the wild-type (h+) viruses can only lyze E. coli B producing mottled or turbid
plaques.
o Occasional T2 mutants appear that break out of their host
cell earlier than normal.
o The mutation occurs in a gene designated r (for "rapid
lysis"). It reveals itself by the extra-large plaques that it forms.
o The wild-type gene, producing a normal time of lysis, is
designated r+. It forms normal-size plaques.
As with
so many organisms, the occurrence of mutations provides the tools to learn
about such things as
- The function of the gene;
Comments
Post a Comment